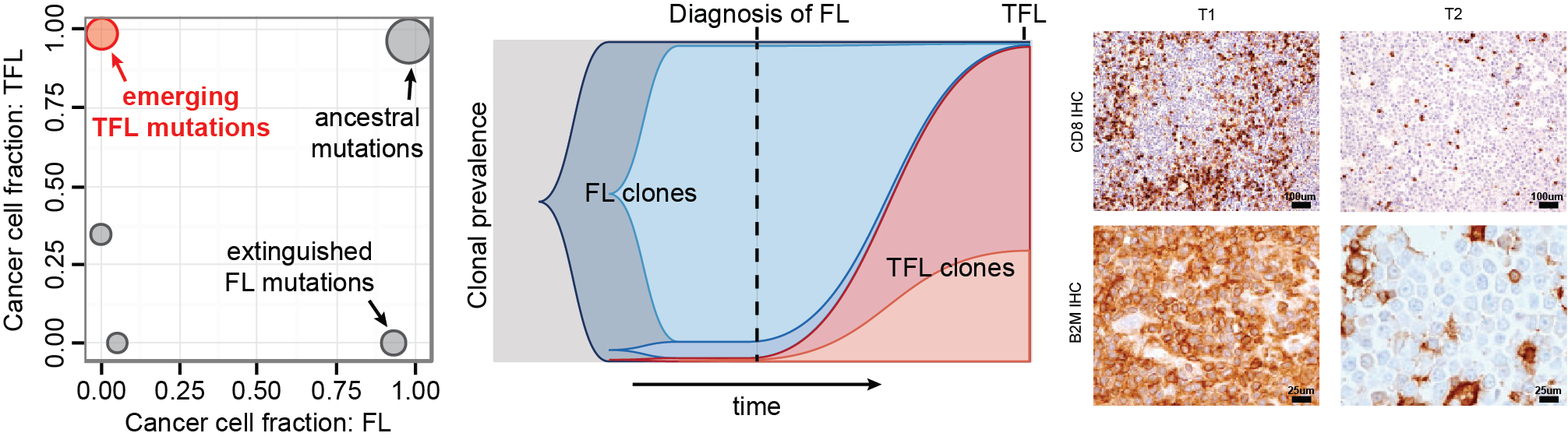
Lymphoma is a biologically diverse disease, and this diversity is reflected in the wide range of outcomes experienced by patients. While some individuals achieve durable remissions, others face early relapse, treatment resistance, or refractory disease. Despite important therapeutic advances, we still lack the ability to consistently tailor treatment to the unique biological vulnerabilities of each patient’s tumour.
Our research program aims to change this. We believe that deeply understanding the molecular, genetic, and immunologic foundations of lymphoma will enable more precise diagnostics and more effective, personalized therapies. By integrating cutting‑edge genomic, epigenomic, and transcriptomic technologies with clinical data, we uncover the mechanisms that drive disease behaviour, tumour evolution, and therapeutic resistance.
A central focus of our program is the development of novel therapeutic strategies, including rational combination approaches and targeted protein degradation platforms designed to exploit tumour‑specific vulnerabilities and overcome resistance. Complementing these efforts, we study circulating tumour DNA as a minimally invasive biomarker for prognosis, risk stratification, treatment adaptation, and real‑time response assessment.
Our ultimate goal is to translate these discoveries into clinically actionable tools—enhancing risk prediction, informing treatment selection, and ultimately improving outcomes for patients with lymphoma.